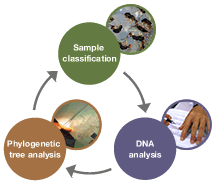
The history of the diversification of living creatures is imprinted in DNA sequences. The phylogenetic tree derived from a comparison of the DNA of every species of the fig and the fig wasp shows the sequence at which these species divide (genetic relationship). The most important part of this work is collecting the samples that will serve as the basis of the research.
The editing work is difficult. When amplifying the DNA areas I want to compare with PCR and determining DNA sequences, the sequences obtained are reconnected using a computer and the portions in which the context cannot be determined are discarded. The DNA sequences arranged in this way is aligned; that is, gaps are inserted and arranged so that the DNA sequences obtained from the different samples are arrayed together. At last, work starts on creating the phylogenetic tree. Finishing the tree does not mean the work is completed, however.
When the phylogenetic tree created by focusing on certain genes has an unavoidably low degree of reliability, it is necessary to recreate the tree using other genes. The volume of the work then increases rather substantially. Therefore, we have to select the genes carefully.
(Zhi-Hui Su) |
 |
 |
|
|